Difference between revisions of "Main Page"
(Add HyMS. Videos not working yet.) |
m |
||
| (3 intermediate revisions by the same user not shown) | |||
| Line 5: | Line 5: | ||
= Hybrid Multiplexed Sculpted Light (HyMS) Microscopy = | = Hybrid Multiplexed Sculpted Light (HyMS) Microscopy = | ||
| − | + | We have developed a modular platform named '''Hybrid Multiplexed Sculpted Light (HyMS) microscopy''' featuring a systems-wide design paradigm that maximizes the acquisition volume size and speed while maintaining fidelity for obtaining single neuron activity traces. Our modular design utilizes a hybrid two- and three-photon acquisition and allows for volumetric recording of neuroactivity at single-cell resolution within up to '''1 × 1 × 1.22 mm volumes at up to 17 Hz''' in awake behaving mice. We have established the capabilities and potential of the different configurations of HyMS microscopy at depth and across brain regions by applying it to ''in-vivo'' recording of '''up to 12,000 neurons''' in mouse auditory cortex, posterior parietal cortex, and hippocampus. | |
| − | We | + | We demonstrated the performance of HyMS microscopy and its different configurations in the mouse posterior parietal cortex, primary auditory cortex, and the hippocampus. |
| − | |||
| − | |||
[[File:fig1-01-1024x588.png|thumb|center|upright=2.0|alt=Figure 1: Different acquisition configurations of the HyMS microscopy.|'''Figure 1: Different acquisition configurations of the HyMS microscopy.''']] | [[File:fig1-01-1024x588.png|thumb|center|upright=2.0|alt=Figure 1: Different acquisition configurations of the HyMS microscopy.|'''Figure 1: Different acquisition configurations of the HyMS microscopy.''']] | ||
| Line 15: | Line 13: | ||
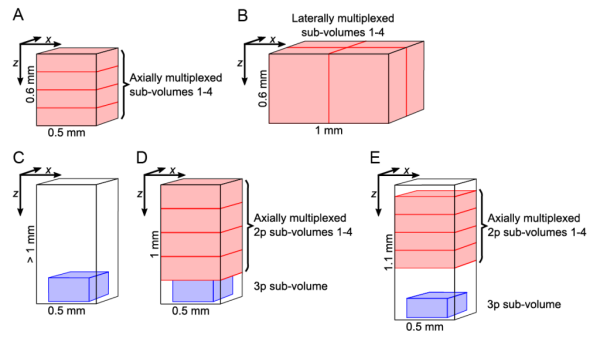
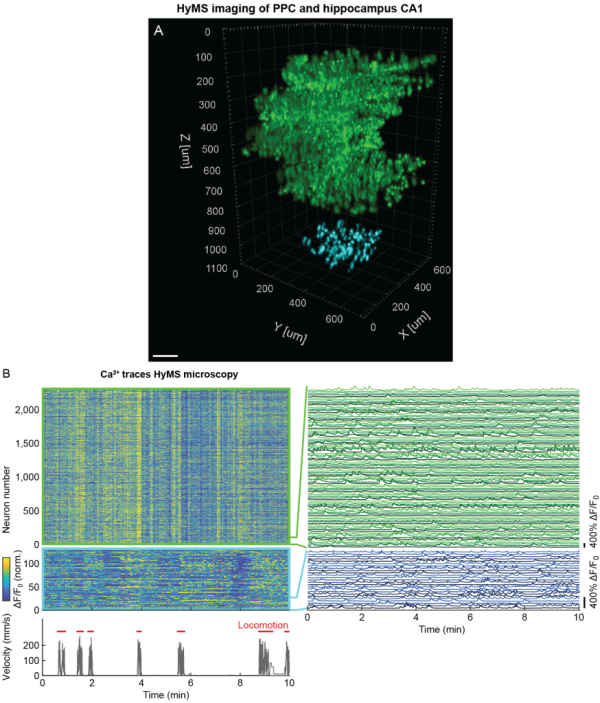
Our target volumes span axially over cortical layers 1–6, encompassing basal dendritic and pyramidal layers of the hippocampus CA1 region, within the hippocampus from CA1 to the dentate gyrus, and cover laterally up to 1 mm<sup>2</sup>. In our different configurations, we show calcium (Ca<sup>2+</sup>) of neurons within volumes of up to 1 × 1 × 0.6 mm at >5 Hz, or 340 × 340 × 250 µm located at ~1 mm depth, or ~0.7 × 0.7 × 1 mm spanning an entire cortical column and a major portion of the hippocampus. Further, we have pushed the attainable functional high-speed imaging depth to >1.2 mm. Our method has enabled near-simultaneous ''in vivo'' recording from up to 12,000 neurons across different regions and depths of the mouse brain. Thereby it opens up new possibilities to study how distributed network dynamics of large neuron populations encode sensory inputs and generate behavior. | Our target volumes span axially over cortical layers 1–6, encompassing basal dendritic and pyramidal layers of the hippocampus CA1 region, within the hippocampus from CA1 to the dentate gyrus, and cover laterally up to 1 mm<sup>2</sup>. In our different configurations, we show calcium (Ca<sup>2+</sup>) of neurons within volumes of up to 1 × 1 × 0.6 mm at >5 Hz, or 340 × 340 × 250 µm located at ~1 mm depth, or ~0.7 × 0.7 × 1 mm spanning an entire cortical column and a major portion of the hippocampus. Further, we have pushed the attainable functional high-speed imaging depth to >1.2 mm. Our method has enabled near-simultaneous ''in vivo'' recording from up to 12,000 neurons across different regions and depths of the mouse brain. Thereby it opens up new possibilities to study how distributed network dynamics of large neuron populations encode sensory inputs and generate behavior. | ||
| + | <!-- | ||
[[File:HyMS_video1.mov|thumb|center|upright=1.5|alt=Video 1: Example 4x axial volumetric 2p-MuST Ca2+ imaging in mouse PPC.|'''Video 1: Example 4x axial volumetric 2p-MuST Ca<sup>2+</sup> imaging in mouse PPC.''' 3D rendering (MIP) of a 30-min recording in mouse PPC, 690 × 675 × 600 µm volumetric field of view at 16.7 Hz, cytosolic GCaMP6f. The video shows the first 3 min of the recording. Playback speed: 3×.]] | [[File:HyMS_video1.mov|thumb|center|upright=1.5|alt=Video 1: Example 4x axial volumetric 2p-MuST Ca2+ imaging in mouse PPC.|'''Video 1: Example 4x axial volumetric 2p-MuST Ca<sup>2+</sup> imaging in mouse PPC.''' 3D rendering (MIP) of a 30-min recording in mouse PPC, 690 × 675 × 600 µm volumetric field of view at 16.7 Hz, cytosolic GCaMP6f. The video shows the first 3 min of the recording. Playback speed: 3×.]] | ||
| − | + | --> | |
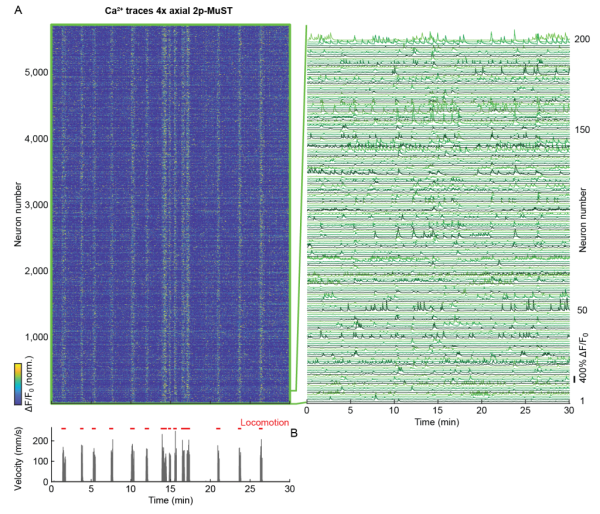
| − | + | In the 4x axial (Multiplexed Scanned Temporal Focusing) MuST configuration, we recorded over a 600 µm axial range with each of the four beamlets covering a 150 µm sub-volume. Using this configuration, we recorded within a volume of ~690 × 675 × 600 µm at 16.7 Hz. Depending on the recording duration (~10–30 min) and the animal’s behavioral state, we typically found ~3,000–6,000 active neurons in our imaging volume. | |
[[File:fig2-01-1024x878.png|thumb|center|upright=2.0|alt=Figure 2: High-speed volumetric 4x-axial 2p-MuST imaging of mouse PPC.|'''Figure 2: High-speed volumetric 4x-axial 2p-MuST imaging of mouse PPC.''']] | [[File:fig2-01-1024x878.png|thumb|center|upright=2.0|alt=Figure 2: High-speed volumetric 4x-axial 2p-MuST imaging of mouse PPC.|'''Figure 2: High-speed volumetric 4x-axial 2p-MuST imaging of mouse PPC.''']] | ||
| − | + | <!-- | |
[[File:HyMS_video2.mov|thumb|center|upright=1.5|alt=Video 2: Example of 4x lateral 2p-MuST recording of mouse primary auditory cortex.|'''Video 2: Example of 4x lateral 2p-MuST recording of mouse primary auditory cortex.''' 3D rendering of a 20-min volumetric 4x-lateral 2p-MuST recording in mouse primary auditory cortex, 1,000 × 1,000 × 600 µm FOV, 5.1 Hz, double-transgenic GCaMP6s. The video shows the first 3 min of the recording. Playback speed: 3×.]] | [[File:HyMS_video2.mov|thumb|center|upright=1.5|alt=Video 2: Example of 4x lateral 2p-MuST recording of mouse primary auditory cortex.|'''Video 2: Example of 4x lateral 2p-MuST recording of mouse primary auditory cortex.''' 3D rendering of a 20-min volumetric 4x-lateral 2p-MuST recording in mouse primary auditory cortex, 1,000 × 1,000 × 600 µm FOV, 5.1 Hz, double-transgenic GCaMP6s. The video shows the first 3 min of the recording. Playback speed: 3×.]] | ||
| − | + | --> | |
Using our optimized 3p imaging configuration allowed us to perform high-speed single-plane and volumetric 3p recording at depths up to 1.22 mm below the brain surface. | Using our optimized 3p imaging configuration allowed us to perform high-speed single-plane and volumetric 3p recording at depths up to 1.22 mm below the brain surface. | ||
| − | + | <!-- | |
[[File:HYMS_video3.mov|thumb|center|upright=1.5|alt=Video 3: Example single-plane 3p recording of mouse hippocampus CA1 through intact cortex.|'''Video 3: Example single-plane 3p recording of mouse hippocampus CA1 through intact cortex.''' Single-plane 3p recording in mouse hippocampus CA1 through intact cortex at 1,160 µm depth, 340 × 340 µm FOV, 78 Hz, cytosolic GCaMP6s. Left: Unprocessed raw dataset (except for motion correction using NoRMCorre). Right: Denoised dataset. Scalebar: 100 µm. The video shows the first 5 min of the recording. Playback speed: 5×.]] | [[File:HYMS_video3.mov|thumb|center|upright=1.5|alt=Video 3: Example single-plane 3p recording of mouse hippocampus CA1 through intact cortex.|'''Video 3: Example single-plane 3p recording of mouse hippocampus CA1 through intact cortex.''' Single-plane 3p recording in mouse hippocampus CA1 through intact cortex at 1,160 µm depth, 340 × 340 µm FOV, 78 Hz, cytosolic GCaMP6s. Left: Unprocessed raw dataset (except for motion correction using NoRMCorre). Right: Denoised dataset. Scalebar: 100 µm. The video shows the first 5 min of the recording. Playback speed: 5×.]] | ||
| − | + | --> | |
We could also demonstrate volumetric Ca<sup>2+</sup> imaging at 750–1,000 µm tissue depths of a 340 × 340 × 250 µm volume at 3.9 Hz. | We could also demonstrate volumetric Ca<sup>2+</sup> imaging at 750–1,000 µm tissue depths of a 340 × 340 × 250 µm volume at 3.9 Hz. | ||
| − | + | <!-- | |
[[File:HYMS_video4.mov|thumb|center|upright=1.5|alt=Video 4: Example volumetric 3p recording of mouse PPC layer 6.|'''Video 4: Example volumetric 3p recording of mouse PPC layer 6.''' 3D rendering of a 5-min volumetric 3p recording in mouse PPC L6 through intact cortex at 750–1,000 µm depth, 340 × 340 × 250 µm FOV, 3.9 Hz, cytosolic GCaMP6f. Playback speed: 5×.]] | [[File:HYMS_video4.mov|thumb|center|upright=1.5|alt=Video 4: Example volumetric 3p recording of mouse PPC layer 6.|'''Video 4: Example volumetric 3p recording of mouse PPC layer 6.''' 3D rendering of a 5-min volumetric 3p recording in mouse PPC L6 through intact cortex at 750–1,000 µm depth, 340 × 340 × 250 µm FOV, 3.9 Hz, cytosolic GCaMP6f. Playback speed: 5×.]] | ||
| − | + | --> | |
HyMS microscopy is capable of accessing brain regions residing below the neocortex such as the hippocampus, overcoming the limitations forced by cortical aspiration of the overlying structures that may have critical inputs or fibers of passage to the target volume. | HyMS microscopy is capable of accessing brain regions residing below the neocortex such as the hippocampus, overcoming the limitations forced by cortical aspiration of the overlying structures that may have critical inputs or fibers of passage to the target volume. | ||
| Line 43: | Line 42: | ||
== Technical documentation == | == Technical documentation == | ||
| − | = Scanned Temporal Focusing ( | + | = Scanned Temporal Focusing (Scan-TeFo) Microscopy = |
| − | Scanned Temporal Focusing ( | + | Scanned Temporal Focusing (Scan-TeFo) is a method based on light sculpting that enables unbiased single and dual-plane high-speed calcium imaging, as well as in vivo volumetric calcium imaging of volumes near the size of a mouse cortical column (500x500x500 µm) at single-cell resolution and fast volume rates (3 - 6 Hz). This is achieved by tailoring the point-spread function of our microscope to the structures of interest while maximizing the signal-to-noise ratio while using a home-built fiber laser amplifier with pulses that are synchronized to the imaging voxel speed. Together, they enable near-simultaneous in-vivo recording of calcium dynamics of several thousand active neurons across cortical layers and in the hippocampus of awake behaving mice. |
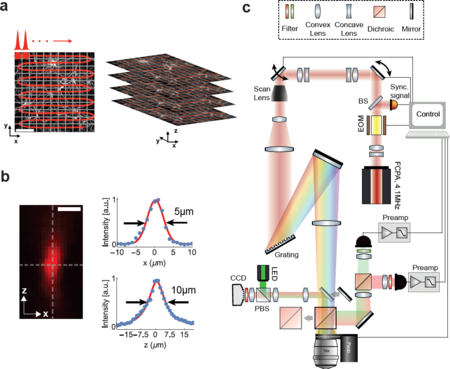
| − | [[File:Fig1.png|thumb|center|upright=1.5|alt=Figure 1: Schematic and principle of scanned temporal focusing imaging system.|'''Figure 1: Schematic and principle of scanned temporal focusing imaging system.''' (a) Schematic of the | + | [[File:Fig1.png|thumb|center|upright=1.5|alt=Figure 1: Schematic and principle of scanned temporal focusing imaging system.|'''Figure 1: Schematic and principle of scanned temporal focusing imaging system.''' (a) Schematic of the Scan-TeFo imaging. A large field-of-view is raster-scanned using an enlarged sculpted PSF and a one-pulse-per voxel excitation-acquisition scheme. Volumetric image acquisition is achieved by translating the objective axially (along z-axis) via a high-speed long-range piezo. Scale bar is 100µm. (b) Measured axial confinement of the sculpted point-spread-function (PSF) of the laterally symmetric TeFo-spot using 0.5µm sized beads. Corresponding x and z profiles showing lateral and axial confinement of excitation, a.u., arbitrary units. Scale bar is 10µm. (c) Overview of the Scan-TeFo microscope. The main dichroic mirror and a mirror are mounted on a slide in order to switch between two-photon scanning and wide-field epi-fluorescent imaging mode. EOM, electro-optical modulator, BS, beam-splitter, PBS, polarizing beam-splitter.]] |
| − | + | Scan-TeFo, combines three key ideas; (1) excite the minimally required number of voxels per unit volume necessary to resolve the structure of interest, (2) ensure a near-isotropic 3D shape of these excitation voxels and (3) maximize the obtainable fluorescence signal-to-noise from each voxel by synchronizing the laser pulse repetition to the voxel acquisition rate. These conditions increase the volume acquisition rate by eliminating the unnecessary oversampling in the lateral (x,y) plane as is some of the applications of conventional 2PM just for obtaining the necessary axial localization of excitation. At the same time, it allows maximizing the obtainable fluorescence signal-to-noise for a given average excitation power. The enlarged sculpted PSF volume in combination with the one pulse per voxel excitation scheme allows for a higher total signal-to-noise ratio from each GCaMP-expressing neuron at the same excitation power density. This is in essence because larger voxels lead to a higher total collected fluorescence from each voxel and also because at a lower total number of voxels a lower repetition rate can be used – while maintaining a one-pulse per pixel excitation condition – from which the signal benefits non-linearly. Thus, for the same obtainable signal to noise ratio we can either reduce the laser pulse energy per unit area and time (J/(µm2/s)) which in turn improves cell viability or the higher obtainable signal-to-noise ratio in case of Scan-TeFo can be used to reduce the voxel dwell time and thereby speed up the voxel acquisition rate. Furthermore, as shown previously, using temporal focusing also provides the additional advantage that the axial confinement of excitation is more resilient to aberrations and tissue scattering compared to conventional two-photon excitation. | |
| − | A schematic of our | + | A schematic of our Scan-TeFo imaging approach is depicted in Fig. 1a along with a measurement of the corresponding 3D PSF (Fig. 1b), and the schematic of the setup (Fig. 1c). The enlarged 3D PSF has about a 130-fold increased excitation volume of ~130 µm3 compared to the PSF in diffraction-limited scanning. This required a corresponding increase in pulse energy in order to maintain a comparable power density (J/(µm2/s)), which in turn necessitated the development of an alternative femtosecond laser that can provide sufficient pulse energies at repetition rates equal to or higher than the image acquisition rate. Whereas a repetition rate higher than the imaging/voxel rate would be possible in principle, the highest signal-to-noise is achieved with a one-pulse-per-voxel excitation approach, as this scheme optimizes the obtainable signal and prevents integration of detector read noise that can accumulate when fluorescence from multiple pulses is integrated. |
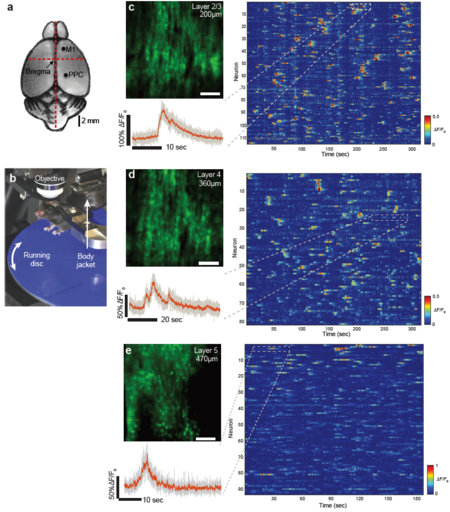
| − | [[File:Fig2.png|thumb|center|upright=1.5|alt=Figure 2: High-speed single plane Ca2+-imaging in mouse posterior parietal cortex at 158fps using | + | [[File:Fig2.png|thumb|center|upright=1.5|alt=Figure 2: High-speed single plane Ca2+-imaging in mouse posterior parietal cortex at 158fps using Scan-TeFo.|'''Figure 2: High-speed single plane Ca2+-imaging in mouse posterior parietal cortex at 158fps using Scan-TeFo.''' (a) Sketch of the adult mouse brain, indicating the injection and imaging sites. M1 primary motor cortex, PPC posterior parietal cortex. Scale bar is 2mm. (b) Photograph of the animal mounted below the imaging objective via a head bar holder on a homebuilt running disc. The animal is additionally supported by a stabilizer attached to a body jacket. (c) Top left: Time averaged intensity projection image at Layer 2/3 (200µm depth) in mouse PPC expressing GCAMP6m. Right: Calcium traces of 114 active neurons identified in this plane. Each row shows the time-series activity of an individual neuron. Color indicates percent fluorescence changes (∆F/F0); scaling is indicated by the color bar on the right. Typical calcium transients consisted of an estimated (30±10)*103 detected photons (~3 photons per digitized count). Bottom left: Zoom-into a single trace (dashed box) is shown. Grey: raw data, red: moving average of 10 time points. (d) Same as in c, but at Layer-4 (depth of 360µm) in the same animal. (e) Same as in c, but at Layer-5 (depth of 470µm) of a GCAMP6f expressing mouse. Scale bar is 100µm in c, d. Representative recordings out of ten data sets.]] |
| − | As a first demonstration of the capabilities of our | + | As a first demonstration of the capabilities of our Scan-TeFo imaging system, we performed two-photon calcium imaging in Layer 2/3 (200 µm depth) and Layer 4 (360 µm depth) of the posterior parietal cortex (PPC) of awake head-fixed mice expressing the GCaMP6m calcium sensor (see Fig. 2). The mice were placed on a home-built mount that includes a running disk designed to minimize the effects of brain movements. The optimized voxel acquisition and excitation scheme of our Scan-TeFo system allowed us to record the activity of neurons at each plane at 158 Hz. We typically recorded between 150 and 200 neurons in our imaging field-of view (FOV, 500x500µm) of which 80-120 showed activity, depending on imaging depth and the animal’s behavior. Despite the high frame rate and low pixel dwell time (~250 ns), we could obtain sufficient signal-to-noise to extract calcium traces from the data with high. Our demonstrated frame rate of ~160 Hz and field-of-view of 500x500 µm, inside scattering brain tissue, outperforms previously demonstrated, unbiased imaging systems based on scanning two-photon microscopy when our obtained FOV, speed and depth (down to 500 µm) are considered collectively. |
[http://www.nature.com/nmeth/journal/v13/n12/fig_tab/nmeth.4040_SV4.html Video 1: Fast volumetric imaging of 3D Ca2+-dynamics across cortical layers in mouse posterior parietal cortex.] | [http://www.nature.com/nmeth/journal/v13/n12/fig_tab/nmeth.4040_SV4.html Video 1: Fast volumetric imaging of 3D Ca2+-dynamics across cortical layers in mouse posterior parietal cortex.] | ||
| Line 60: | Line 59: | ||
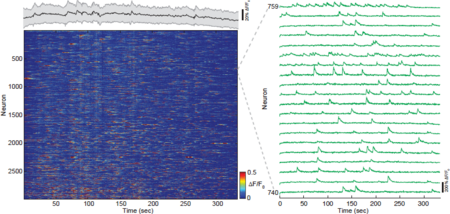
[[File:Fig3.png|thumb|center|upright=1.5|alt=Figure 3: Fast volumetric imaging of Ca2+-dynamics across cortical layers in mouse posterior parietal cortex.|'''Figure 3: Fast volumetric imaging of Ca2+-dynamics across cortical layers in mouse posterior parietal cortex.''' Time-series heat map of all active neurons (2826 from a total of 4306) during a ~330sec recording from a 500x500x500µm volume in the cortex (PPC) in an awake mouse. Volume acquisition rate is 3Hz. Zoom-in of individual traces shows distinct calcium transients (right panel). Graph on top of the heat map shows average fluorescence level of all neurons during the recording (black line) as well as s.e.m (grey shading), indicating negligible photo-bleaching. ]] | [[File:Fig3.png|thumb|center|upright=1.5|alt=Figure 3: Fast volumetric imaging of Ca2+-dynamics across cortical layers in mouse posterior parietal cortex.|'''Figure 3: Fast volumetric imaging of Ca2+-dynamics across cortical layers in mouse posterior parietal cortex.''' Time-series heat map of all active neurons (2826 from a total of 4306) during a ~330sec recording from a 500x500x500µm volume in the cortex (PPC) in an awake mouse. Volume acquisition rate is 3Hz. Zoom-in of individual traces shows distinct calcium transients (right panel). Graph on top of the heat map shows average fluorescence level of all neurons during the recording (black line) as well as s.e.m (grey shading), indicating negligible photo-bleaching. ]] | ||
| − | To demonstrate the full potential of our | + | To demonstrate the full potential of our Scan-TeFo method for fast and unbiased, three-dimensional imaging, we performed continuous volumetric Ca2+-imaging over a V-FOV of 500x500x500 µm in the PPC of awake behaving mice at a volume rate of 3 Hz (Video 1 and Fig. 3). This volume covers a major portion of a cortical column, and includes layers I-V in the mouse PPC cortex 52. Most individual Ca2+-traces show pronounced maximum fluorescence changes (∆F/F0), exemplifying the quality and signal-to-noise of the recording. In our data sets we typically identified several thousand (~2000-4000) neurons within our recording volumes, out of which a large portion (~60 %) showed activity during a typical recording time duration of ~ 5 min. |
== Reference == | == Reference == | ||
Latest revision as of 19:13, 20 December 2024
Contents
Welcome to the Scan-TeFo & HyMS wiki
This wiki contains models, parts lists, assembly instructions and other documentation that allows to build and set up our Scanned Temporal Focusing (Scan-TeFo) Microscope Prevedel et al., Nature Methods (2016), as well as our more recent and advanced Hybrid Multiplexed Sculpted Light (HyMS) Microscope Weisenburger et al., Cell (2019).
In what follows, we summarize the Scan-TeFo and HyMS approaches and provide links to example datasets and technical documentation.
Hybrid Multiplexed Sculpted Light (HyMS) Microscopy
We have developed a modular platform named Hybrid Multiplexed Sculpted Light (HyMS) microscopy featuring a systems-wide design paradigm that maximizes the acquisition volume size and speed while maintaining fidelity for obtaining single neuron activity traces. Our modular design utilizes a hybrid two- and three-photon acquisition and allows for volumetric recording of neuroactivity at single-cell resolution within up to 1 × 1 × 1.22 mm volumes at up to 17 Hz in awake behaving mice. We have established the capabilities and potential of the different configurations of HyMS microscopy at depth and across brain regions by applying it to in-vivo recording of up to 12,000 neurons in mouse auditory cortex, posterior parietal cortex, and hippocampus.
We demonstrated the performance of HyMS microscopy and its different configurations in the mouse posterior parietal cortex, primary auditory cortex, and the hippocampus.
Our target volumes span axially over cortical layers 1–6, encompassing basal dendritic and pyramidal layers of the hippocampus CA1 region, within the hippocampus from CA1 to the dentate gyrus, and cover laterally up to 1 mm2. In our different configurations, we show calcium (Ca2+) of neurons within volumes of up to 1 × 1 × 0.6 mm at >5 Hz, or 340 × 340 × 250 µm located at ~1 mm depth, or ~0.7 × 0.7 × 1 mm spanning an entire cortical column and a major portion of the hippocampus. Further, we have pushed the attainable functional high-speed imaging depth to >1.2 mm. Our method has enabled near-simultaneous in vivo recording from up to 12,000 neurons across different regions and depths of the mouse brain. Thereby it opens up new possibilities to study how distributed network dynamics of large neuron populations encode sensory inputs and generate behavior.
In the 4x axial (Multiplexed Scanned Temporal Focusing) MuST configuration, we recorded over a 600 µm axial range with each of the four beamlets covering a 150 µm sub-volume. Using this configuration, we recorded within a volume of ~690 × 675 × 600 µm at 16.7 Hz. Depending on the recording duration (~10–30 min) and the animal’s behavioral state, we typically found ~3,000–6,000 active neurons in our imaging volume.
Using our optimized 3p imaging configuration allowed us to perform high-speed single-plane and volumetric 3p recording at depths up to 1.22 mm below the brain surface. We could also demonstrate volumetric Ca2+ imaging at 750–1,000 µm tissue depths of a 340 × 340 × 250 µm volume at 3.9 Hz. HyMS microscopy is capable of accessing brain regions residing below the neocortex such as the hippocampus, overcoming the limitations forced by cortical aspiration of the overlying structures that may have critical inputs or fibers of passage to the target volume.
Reference
Siegfried Weisenburger, Frank Tejera, Jeffrey Demas, Brandon Chen, Jason Manley, Fraser T. Sparks, Francisca Martínez Traub, Tanya Daigle, Hongkui Zeng, Attila Losonczy, and Alipasha Vaziri Volumetric Ca2+ Imaging in the Mouse Brain using Hybrid Multiplexed Sculpted Light (HyMS) Microscopy Cell 177, 1-17 (2019). (Download)
Technical documentation
Scanned Temporal Focusing (Scan-TeFo) Microscopy
Scanned Temporal Focusing (Scan-TeFo) is a method based on light sculpting that enables unbiased single and dual-plane high-speed calcium imaging, as well as in vivo volumetric calcium imaging of volumes near the size of a mouse cortical column (500x500x500 µm) at single-cell resolution and fast volume rates (3 - 6 Hz). This is achieved by tailoring the point-spread function of our microscope to the structures of interest while maximizing the signal-to-noise ratio while using a home-built fiber laser amplifier with pulses that are synchronized to the imaging voxel speed. Together, they enable near-simultaneous in-vivo recording of calcium dynamics of several thousand active neurons across cortical layers and in the hippocampus of awake behaving mice.
Scan-TeFo, combines three key ideas; (1) excite the minimally required number of voxels per unit volume necessary to resolve the structure of interest, (2) ensure a near-isotropic 3D shape of these excitation voxels and (3) maximize the obtainable fluorescence signal-to-noise from each voxel by synchronizing the laser pulse repetition to the voxel acquisition rate. These conditions increase the volume acquisition rate by eliminating the unnecessary oversampling in the lateral (x,y) plane as is some of the applications of conventional 2PM just for obtaining the necessary axial localization of excitation. At the same time, it allows maximizing the obtainable fluorescence signal-to-noise for a given average excitation power. The enlarged sculpted PSF volume in combination with the one pulse per voxel excitation scheme allows for a higher total signal-to-noise ratio from each GCaMP-expressing neuron at the same excitation power density. This is in essence because larger voxels lead to a higher total collected fluorescence from each voxel and also because at a lower total number of voxels a lower repetition rate can be used – while maintaining a one-pulse per pixel excitation condition – from which the signal benefits non-linearly. Thus, for the same obtainable signal to noise ratio we can either reduce the laser pulse energy per unit area and time (J/(µm2/s)) which in turn improves cell viability or the higher obtainable signal-to-noise ratio in case of Scan-TeFo can be used to reduce the voxel dwell time and thereby speed up the voxel acquisition rate. Furthermore, as shown previously, using temporal focusing also provides the additional advantage that the axial confinement of excitation is more resilient to aberrations and tissue scattering compared to conventional two-photon excitation.
A schematic of our Scan-TeFo imaging approach is depicted in Fig. 1a along with a measurement of the corresponding 3D PSF (Fig. 1b), and the schematic of the setup (Fig. 1c). The enlarged 3D PSF has about a 130-fold increased excitation volume of ~130 µm3 compared to the PSF in diffraction-limited scanning. This required a corresponding increase in pulse energy in order to maintain a comparable power density (J/(µm2/s)), which in turn necessitated the development of an alternative femtosecond laser that can provide sufficient pulse energies at repetition rates equal to or higher than the image acquisition rate. Whereas a repetition rate higher than the imaging/voxel rate would be possible in principle, the highest signal-to-noise is achieved with a one-pulse-per-voxel excitation approach, as this scheme optimizes the obtainable signal and prevents integration of detector read noise that can accumulate when fluorescence from multiple pulses is integrated.
As a first demonstration of the capabilities of our Scan-TeFo imaging system, we performed two-photon calcium imaging in Layer 2/3 (200 µm depth) and Layer 4 (360 µm depth) of the posterior parietal cortex (PPC) of awake head-fixed mice expressing the GCaMP6m calcium sensor (see Fig. 2). The mice were placed on a home-built mount that includes a running disk designed to minimize the effects of brain movements. The optimized voxel acquisition and excitation scheme of our Scan-TeFo system allowed us to record the activity of neurons at each plane at 158 Hz. We typically recorded between 150 and 200 neurons in our imaging field-of view (FOV, 500x500µm) of which 80-120 showed activity, depending on imaging depth and the animal’s behavior. Despite the high frame rate and low pixel dwell time (~250 ns), we could obtain sufficient signal-to-noise to extract calcium traces from the data with high. Our demonstrated frame rate of ~160 Hz and field-of-view of 500x500 µm, inside scattering brain tissue, outperforms previously demonstrated, unbiased imaging systems based on scanning two-photon microscopy when our obtained FOV, speed and depth (down to 500 µm) are considered collectively.
To demonstrate the full potential of our Scan-TeFo method for fast and unbiased, three-dimensional imaging, we performed continuous volumetric Ca2+-imaging over a V-FOV of 500x500x500 µm in the PPC of awake behaving mice at a volume rate of 3 Hz (Video 1 and Fig. 3). This volume covers a major portion of a cortical column, and includes layers I-V in the mouse PPC cortex 52. Most individual Ca2+-traces show pronounced maximum fluorescence changes (∆F/F0), exemplifying the quality and signal-to-noise of the recording. In our data sets we typically identified several thousand (~2000-4000) neurons within our recording volumes, out of which a large portion (~60 %) showed activity during a typical recording time duration of ~ 5 min.
Reference
Robert Prevedel, Aart J. Verhoef, Alejandro J. Pernia-Andrade*, Siegfried Weisenburger*, Ben S. Huang, Tobias Nöbauer, Alma Fernández, Jeroen E. Delcour, Peyman Golshani, Andrius Baltuska and Alipasha Vaziri, "Fast volumetric calcium imaging across multiple cortical layers using sculpted light", Nature Methods 13, 1021 – 1028 (2016). (Download)